Screening the binding affinity of bile acid derivatives for the glucocorticoid receptor ligand-binding domain Scientific paper
Main Article Content
Abstract
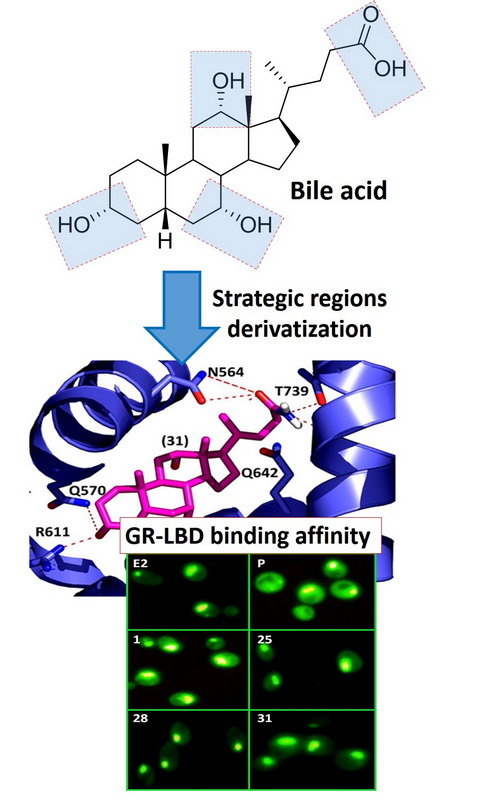
The necessity of anti-inflammatory drugs such as glucocorticoids has been evident during the COVID-19 pandemic. Glucocorticoids, are the standard therapy for the treatment of moderate and severe COVID-19 patients. However, serious side effects limit the use of these drugs, and anti-inflammatory drugs with better pharmacological properties are urgently required. Bile acids are of interest, because of their anti-inflammatory and immunomodulatory properties, facilitated through an unclear mechanism involving transmembrane and nuclear receptors. In this work, we screened the binding activity of a number of bile acid derivatives, for the ligand-binding domain of glucocorticoid receptor (GR-LBD), the most important receptor for anti-inflammatory processes. Tested compounds include oximes, lactones, lactams, tetrazoles, dienones, C-24 alcohols and cholic acid amides. Cholic acid oxime, deoxycholic acid dienone, 3-keto-24-cholic alcohol and cholic acid amide showed best binding affinities for GR-LBD among tested compounds. The in silico molecular docking explanation is provided. SAR analysis showed that expansion of B and C steroid rings or attachment of heterocycle to C ring is not beneficial for binding; side chain should contain hydrogen donor group; the GR-LBD tolerate well different functionalities on C-3 position. These results provide valuable information toward synthesis of the new glucocorticoids based on bile acids.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Ministarstvo Prosvete, Nauke i Tehnološkog Razvoja
Grant numbers 451-03-9/2021-14/ 200125
References
N. Sundahl, J. Bridelance, C. Libert, K. De Bosscher, I. M. Beck, Pharmacol. Ther. 152 (2015) 28 (https://doi.org/10.1016/j.pharmthera.2015.05.001)
F. Buttgereit, R. H. Straub, M. Wehling, G. R. Burmester, Arthritis Rheumatol. 50 (2004) 50 3408 (https://doi.org/10.1002/art.20583)
RECOVERY collaborative group*, N. Engl. J. Med. 384 (2021) 693 (https://doi.org/10.1056/NEJMoa2021436)
J. Souffriau, M. Eggermont, S. Van Ryckeghem, K. Van Looveren, L. Van Wyngene, E. Van Hamme, M. Vuylsteke, R. Beyaert, K. De Bosscher, C. Libert, Sci. Rep. 8 (2018) 12894. (https://doi.org/10.1038/s41598-018-31150-w)
J. Vandewalle, A. Luypaert, K. De Bosscher, C. Libert. Trends Endocrinol. Metab. 29 (2018) 42 (https://doi.org/10.1016/j.tem.2017.10.010)
A. Louw. Front. Immunol. 10 (2019) 1693 (https://doi.org/10.3389/fimmu.2019.01693)
E. Lontchi-Yimagou, E. Sobngwi, T. E. Matsha, A. P. Kengne. Curr. Diab. Rep. 13 (2013) 435 (https://doi.org/10.1007/s11892-013-0375-y)
P. S. Hench. Br. Med. J. 20 (1938) 394 (https://doi.org/10.1136/bmj.2.4050.394)
The Nobel Prize, https://www.nobelprize.org/prizes/medicine/1950/summary/ (20.06.2022.)
R. M. Gadaleta, M. Cariello, C. Sabbà, A. Moschetta, Biochim. Biophys. Acta 1851 (2015) 30 (https://doi.org/10.1016/j.bbalip.2014.08.005)
J. Hageman, H. Herrema, A. K. Groen, F. Kuipers, Arterioscler. Thromb. Vasc. Biol. 30 (2010) 1519 (https://doi.org/10.1161/ATVBAHA.109.197897)
A. Perino, K. Schoonjans, Trends. Pharmacol. Sci. 12 (2015) 847 (https://doi.org/10.1016/j.tips.2015.08.002)
B. Vasiljević, E. Petri, S. Bekić, A Ćelić, Lj. Grbović, K. Pavlović, RSC Med. Chem. 12 (2021) 278 (https://doi.org/10.1039/D0MD00311E)
L. Li, C. Liu, W. Mao, B. Tumen, P. Li, Molecules 24 (2019) 4513. (https://doi.org/10.3390/molecules24244513)
T. Takigawa, H. Miyazaki, M. Kinoshita, N. Kawarabayashi, K. Nishiyama, K. Hatsuse, S. Ono, D. Saitoh, S. Seki, J. Yamamoto, Am. J. Physiol. Gastrointest. Liver Physiol. 305 (2013) G427 (https://doi.org/10.1152/ajpgi.00205.2012)
M. Poša, S. Bjedov, V. Tepavčević, M. Mikulić, M. Sakač, J. Mol. Liq. 303 (2020) 112634 (https://doi.org/10.1016/j.molliq.2020.112634)
M. N. Iqbal, W. H. Elliott, Steroids 53 (1989) 413 (https://doi.org/10.1016/0039-128X(89)90022-6)
R. Leppik, Steroids 41 (1983) 475 (https://doi.org/10.1016/0039-128X(83)90087-9)
R. Hüttenrauch, Arch. Pharm. Pharm. Med. Chem. 294 (1961) 366 (https://doi.org/10.1002/ardp.19612940608)
K. Tamaki, J. Biochem. 45 (1958) 299 (https://doi.org/10.1093/oxfordjournals.jbchem.a126869)
S. Bekić, M. Marinović, E. Petri, M. Sakač, A. Nikolić, V. Kojić, A. Ćelić, Steroids 130 (2018) 22 (https://doi.org/10.1016/j.steroids.2017.12.002)
S. Muddana, B. Peterson, Chembiochem. 4 (2003) 848 (https://doi.org/10.1002/cbic.200300606)
D. Gietz, A. St Jean, R. A. Woods, R. H. Schiestl, Nucleic. Acids. Res. 20 (1992) 1425. (https://doi.org/10.1093/nar/20.6.1425)
S. Dallakyan, A. J. Olson, Methods. Mol. Biol. 1263 (2015) 243 (https://doi.org/10.1007/978-1-4939-2269-7_19)
M. D. Hanwell, D. E. Curtis, D. C. Lonie, T. Vandermeerschd, E. Zurek, G. R. Hutchison, J. Cheminform. 17 (2012) (https://doi.org/10.1186/1758-2946-4-17)
R. K. Bledsoe, V. G. Montana, T. B. Stanley, C. J. Delves, C. J. Apolito, D. D. McKee, T. G. Consler, D. J. Parks, E. L. Stewart, T. M. Willson, M. H. Lambert, J. T. Moore, K. H. Pearce, H. E. Xu, Cell 110 (2002) 93 (https://doi.org/10.1016/s0092-8674(02)00817-6)
A. Pedretti, A. Mazzolari, S. Gervasoni, L. Fumagalli, G. Vistoli, Bioinformatics 37 (2021) 1174 (https://doi.org/10.1093/bioinformatics/btaa774)
PyMOL, http://www.pymol.org/pymol (15.09.2021.)
N. Meanwell, H. Roth, E. Smith, D. Wedding, and J. Kim Wright, J. Org. Chem. 56 (1991) 6897 (https://doi.org/10.1021/jo00024a036)
Y. Huang, J. Cui, S. Chen, C. Gan, Q. Yao, Q. Lin, Bioorg. Med. Chem. Lett. 23 (2013) 2265 (https://doi.org/10.1016/j.bmcl.2012.08.064)
H. H. Abdu-Allah, T. T. Chang, W. S. Li, Steroids 112 (2016) 54 (https://doi.org/10.1016/j.steroids.2016.04.013)
I. S. Zharinova, A. A. Bilyalova, S. I. Bezzubov, Acta Crystallogr., E 74 (2018) 816 (https://doi.org/10.1107/S2056989018007259)
M. I. Duran, C. González, A. Acosta, A. F. Olea, K. Díaz, L. Espinoza, Int. J. Mol. Sci. 8 (2017) 516 (https://doi.org/10.3390/ijms18030516)
M. Poša, V. Tepavčević, Lj. Grbović, M. Mikulić, K. Pavlović, J. Phys. Org. Chem. 34 (2021) e4133 (https://doi.org/10.1002/poc.4133)
D. Škorić, O. Klisurić, S. Jakimov, M. Sakač, J. Csanádi, Beilstein J. Org. Chem. 17 (2021) 2611 (https://doi.org/10.3762/bjoc.17.174)
D. Milijkovic, K. Kuhajda, J. Hranisavljevic, J. Chem. Res. 2 (1996) 106 (https://open.uns.ac.rs/handle/123456789/12941)
G. M. Morris, R. Huey, W. Lindstrom, M. Sanner, R. Belew, D. Goodsell, A. Olson, J. Comput. Chem. 16 (2009) 2785 (https://doi.org/10.1002/jcc.21256)
T. Mitić, S. Shave, N. Semjonous, I. McNae, D. Cobice, G. Lavarey, S. Webster, P. Hadkoe, B. Walker, R. Andrew, Biochem. Pharmacol. 86 (2013) 146 (https://doi.org/10.1016/j.bcp.2013.02.002)
U. Lind, P. Greenidge, M. Gillner, K. F. Koehler, A. Wright, J. Carlstedt-Duke, J. Biol. Chem. 275 (2000) 19041 (https://doi.org/10.1074/jbc.M000228200).