Development of 2D and 3D QSAR models of pyrazole derivatives as acetylcholine esterase inhibitors Scientific paper
Main Article Content
Abstract
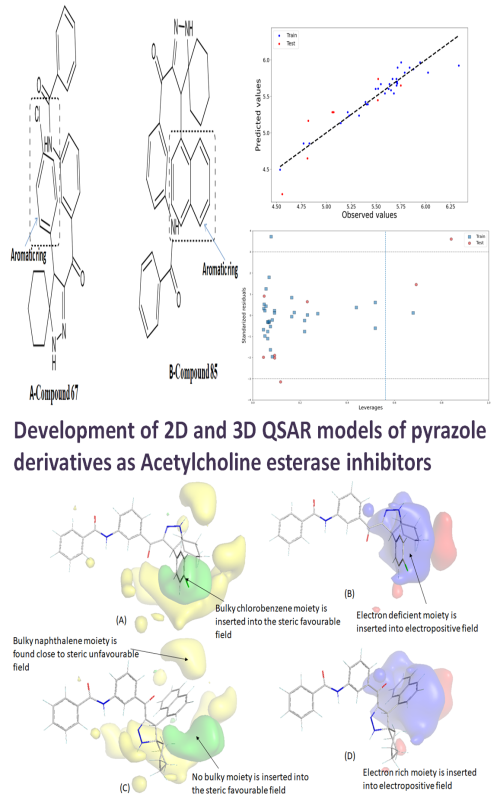
The drugs that are the most useful in all stages of Alzheimer’s disease (AD) are acetylcholinesterase (AChE) inhibitors. The objectives of this work are to generate various QSAR models for such drugs and to select a robust predictive models from the corresponding models. Studies were then focused on finding a range of pyrazole-like AChE inhibitors by 2D and 3D QSAR analysis. The genetic algorithm-based multiple linear regression (GA-MLR) provided the statistically robust 2D QSAR model that depicted the significance of the molecular volume and the number of multiple bonds along with the presence/absence of specific atom-centred fragments and topological distance between 2D pharmacophoric features. Furthermore, these results were correlated well with the electrostatic and steric contour maps retrieved from the 3D QSAR (i.e., alignment-dependent molecular field analysis). The 2D QSAR analysis developed a highly statistical and reliable model, which was compared with the mechanistic interpretation of 3D structures and their electrostatic and steric field contributions leading to a predictive 3D QSAR model. The molecule-protein interactions, elicited by molecular docking, corroborated with the field interactions, as revealed by 2D QSAR. Thus, the developed computational models and simulation analyses in the current work provide valuable information for the future design of pyrazole and spiropyrazoline analogs, as potent AChE inhibitors.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
References
M. Hernández-Rodríguez, J. Correa-Basurto, F. Martínez-Ramos, I. I. Padilla-Martínez, C. G. Benítez-Cardoza, E. Mera-Jiménez, M. C. Rosales-Hernández, J. Alzheimer’s Dis. 41 (2014) 1073 (https://doi.org/10.3233/JAD-140471)
S. Habtemariam, Molecules 24 (2019) 1519 (https://doi.org/10.3390/molecules24081519)
Y. Yamaguchi, H. Miyashita, H. Tsunekawa, A. Mouri, H. C. Kim, K. Saito, T. Matsuno, S. Kawashima, T. Nabeshima, J. Pharmacol. Exp. Ther. 317 (2006) 1079 (https://doi.org/10.1124/jpet.105.098640)
P. Mishra, S. Basak, A. Mukherjee, A. Basu, Lett. Drug Des. Discov. 19 (2021) 192 (https://doi.org/10.2174/1570180818666210813120444)
L. Lecanu, V. Papadopoulos, Recent Pat. CNS Drug Discov. 2 (2007) 113 (https://doi.org/10.2174/157488907780832715)
R. Chawla, A. Arora, M. K. Parameswaran, P. Chan, D. Sharma, S. Michael, T. K. Ravi, Acta Pol. Pharm. 67 (2010) 247 (https://www.ptfarm.pl/pub/File/acta_pol_2010/3_2010/247-253.pdf)
G. Gutti, D. Kumar, P. Paliwal, A. Ganeshpurkar, K. Lahre, A. Kumar, S. Krishnamurthy, S. K. Singh, Bioorg. Chem. 90 (2019) 103080 (https://doi.org/10.1016/j.bioorg.2019.103080)
Ecotoxicological QSARs, K. Roy Ed., Springer 2020, ISBN 978-1071601495
I. Sushko, A. K. Pandey, S. Novotarskyi, R. Körner, M. Rupp, W. Teetz, S. Brandmaier, A. Abdelaziz, V. V. Prokopenko, V. Y. Tanchuk, R. Todeschini, A. Varnek, G. Marcou, P. Ertl, V. Potemkin, M. Grishina, J. Gasteiger, I. I. Baskin, V. A. Palyulin, E. V. Rad-chenko, W. J. Welsh, V. Kholodovych, D. Chekmarev, A. Cherkasov, J. Aires-De-Sousa, Q. Y. Zhang, A. Bender, F. Nigsch, L. Patiny, A. Williams, V. Tkachenko, I. V. Tetko, J. Cheminform. 3 (Suppl. 1) (2011) P20 (https://doi.org/10.1186/1758-2946-3-S1-P20)
P. Ambure, R. B. Aher, A. Gajewicz, T. Puzyn, K. Roy, Chemom. Intell. Lab. Syst. 147 (2015) 1 (https://doi.org/10.1016/j.chemolab.2015.07.007)
P. Tosco, T. Balle, J. Mol. Model. 17 (2011) 201 (https://doi.org/10.1007/s00894-010-0684-x)
J. Sadowski, J. Gasteiger, G. Klebe, J. Chem. Inf. Comp. Sci. 54 (1994) 1000 (https://pubs.acs.org/doi/10.1021/ci00020a039)
A. K. Halder, M. N. D. S. Cordeiro, Biomolecules 11 (2021) 1670 (https://doi.org/10.3390/biom11111670)
G. Kryger, I. Silman, J. L. Sussman, Structure 7 (1999) 297 (https://doi.org/10.1016/S0969-2126(99)80040-9)
H. Safarizadeh, Z. Garkani-Nejad, J. Mol. Graph. Model. 87 (2019) 129 (https://doi.org/10.1016/j.jmgm.2018.11.019)
T. K. Shameera Ahamed, V. K. Rajan, K. Muraleedharan, Food Sci. Hum. Wellness 8 (2019) 53 (https://doi.org/10.1016/j.fshw.2019.02.001)
O. Trott, A. J. Olson, J. Comput. Chem. 31 (2009) 455 (https://doi.org/10.1002/jcc.21334)
G. M. Morris, H. Ruth, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell, A. J. Olson, J. Comput. Chem. 30 (2009) 2785 (https://doi.org/10.1002/jcc.21256)
H. Sugimoto, Pure Appl. Chem. 71 (2009) 2031 (https://doi.org/10.1351/pac199971112031)
P. Mishra, P. Sharma, P. N. Tripathi, S. K. Gupta, P. Srivastava, A. Seth, A. Tripathi, S. Krishnamurthy, S. K. Shrivastava, Bioorg. Chem. 89 (2019) 103025 (https://doi.org/10.1016/j.bioorg.2019.103025)