A DFT study of the chemical bonding properties, aromaticity indexes and molecular docking study of some phenylureas herbicides
Main Article Content
Abstract
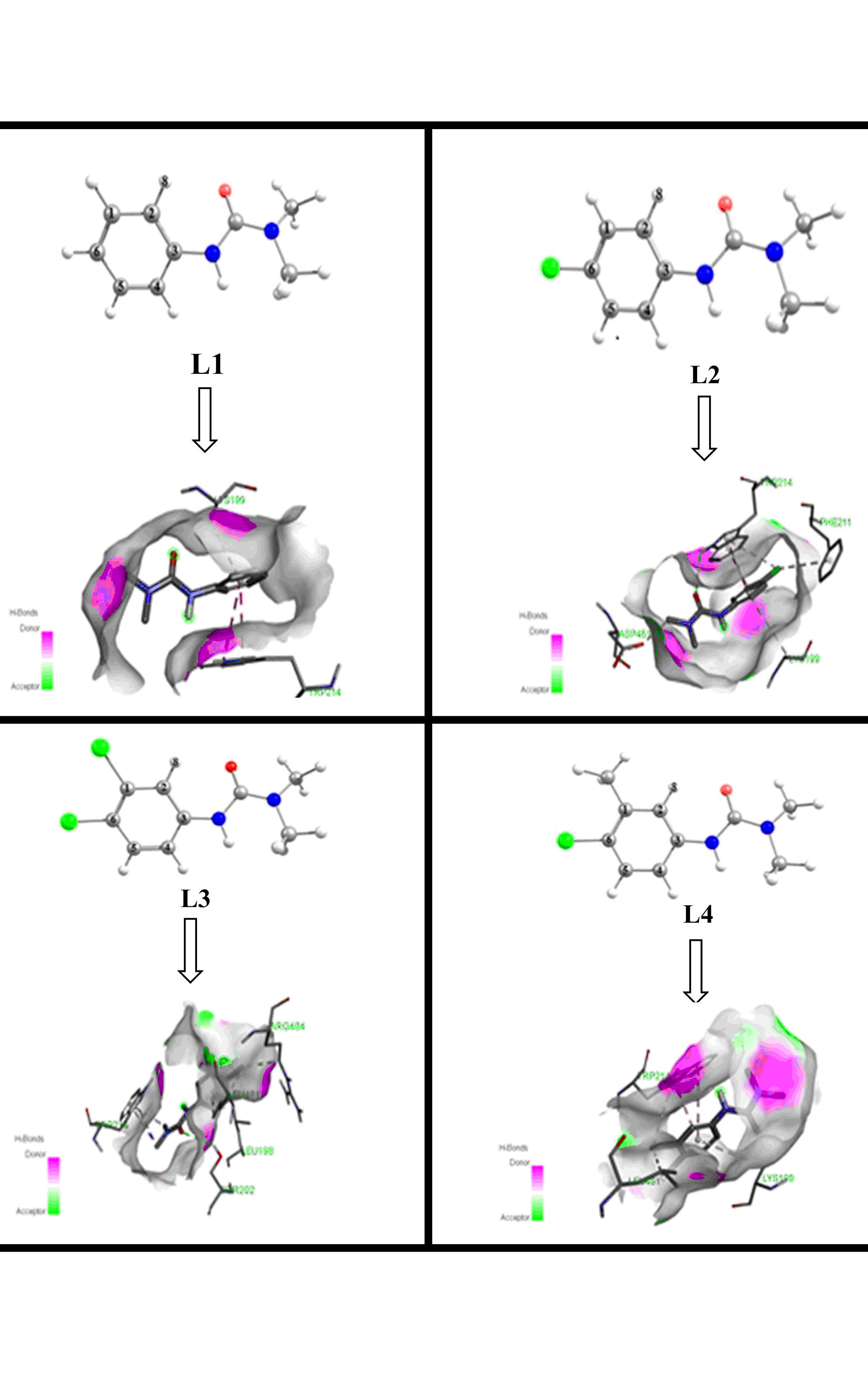
Herbicides have implied disastrous consequences towards the environment and human health. This practice urges scientists to investigate the physical, chemical and biological properties of these substances, hence avoiding the use of the most harmful pesticides. For this purpose, the molecular structure and chemical bonding properties of phenylurea herbicides namely: Fenuron (L1), Monuron (L2), Diuron (L3) and Chlorotoluron (L4), were calculated in water using density functional theory (DFT). The energy decomposition analysis (EDA) and the extended transition state natural orbitals for chemical Valence (ETS-NOCV) reveal the dominant ionic character in Carbon-Nitrogen bond between dimethylurea fragment and benzene ring. Besides, the interaction of these herbicides with the Human Serum Albumin (HSA) was undertaken by molecular modeling. The calculation of HOMA and FLU indexes indicate that the electronic delocalization is stronger in Diuron than the other compounds, mainly caused by the two chloro substituents effects on benzene. Good correlations are found between the calculated parameters such as structural parameters, Mulliken atomic charge, topological and bonding properties and aromaticity indexes. The Vinardo molecular docking results suggest that, the binding energies of the complexes formed between HSA target and investigated compounds have the following order: L3 (-6.59 kcal/mol) < L2 (-6.11 kcal/mol) < L4 (-5.96 kcal/mol) < L1 (-5.76 kcal/mol), which confirmed that the Fenuron is the less harmful option between the studied herbicides especially against HSA.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Ministère de l'Enseignement Supérieur et de la Recherche Scientifique
Grant numbers D01N01UN050220220003
References
P. N. Stamati, S. Maipas, C. Kotampasi, P. Stamatis and L. Hens, Frontiers in Public Health. 4 (2016) 1 (https://doi.org/10.3389/fpubh.2016.00148)
H. Mountacer, L. Tajeddine and M. Sarakha, Herbicides and Environment, Intech, Rijeka, Croatia, 2011 (ISBN 978-953-307-476-4)
R. Kebeish, E. Azab, C. Peterhaensel, R. El-Basheer, Environ. Sci. Pollut. Res. Int. 21 (2014) 8224 (10.1007/s11356-014-2710-5)
J. Jinhoon, K. Sanjida, M. Youngkook, S. Sooim and O.R. Lee, Appl. Biol. Chem. 63 (2020) 1 (https://doi.org/10.1186/s13765-020-00498-x)
T. Vrabelj and M. Finšgar, Biosensors. 12 (2022) 1 (https://doi.org/10.3390/bios12050263)
F. J. Benitez, C. Garcia, J. L. Acero, and F. J. Real, World Academy of Science, Engineering and Technology. 34 (2009) 673 (doi.org/10.5281/zenodo.1078350)
A. Bautista, J. J. Aaron, M. C. Mahedero and A. Muñoz de la Peña, Analysis. 27 (1999) 857 (https://doi.org/10.1051/analusis:1999154)
V. Mile, I. Harsányi, K. Kovács, T. Földes, E. Takács, L. Wojnárovits, Radiation Physics and Chemistry. 132 (2017) 16 (10.1016/j.radphyschem.2016.11.003)
H. Chen, H. Rao, J. Yang J et al, J. Environ. Sci. Health. B 51 (2016) 154 (https://doi.org/10.1080/03601234.2015.1108800)
K. Haruna, S. Veena, Y. Kumar, S. A. Sheena Mary, P. R. Thomas, M. S. Roxy, A. A. Al-Saadi, Heliyon. 5 (2019) 1 (10.1016/j.heliyon.2019.e01987)
L. Humberto M. Huizar, Journal of Chemistry. (2015) 1 (https://doi.org/10.1155/2015/751527)
F. Zhang, B. Liu, G. Liu, Y. Zhang, J. Wang & S. Wang, SCIENTIFIC ReportS. 8 (2018) 1 (https://doi.org/10.1038/s41598-018-21394-x)
A. Andrey, A. Buglak, V. Zherdev, H.T. Lei3, B. Boris, Dzantiev, PLOS ONE. 14 (2019) 1 (https://doi.org/10.1371/journal.pone.0214879)
M. J. Frisch et al, Gaussian 09, Gaussian Inc, Wallingford CT, 2009.
A. D. Becke, The Journal of Chemical Physics. 98 (1933) 5648 (https://doi.org/10.1063/1.464913)
A. D. Becke, Physical Review. A 38 (1988) 3098 (https://doi.org/10.1103/PhysRevA.38.3098)
C. Lee, W. Yang, and R. G. Parr, Physical Review. B 37 (1988) 785 (https://doi.org/10.1103/PhysRevB.37.785)
F. M. Bickelhaupt, E. J. Baerends, Reviews in computational chemistry, K. B. Lipkowitz and D. B, Boyd, Eds., Wiley-VCH, New York, 2000, pp. 1-86 (https://doi.org/10.1002/9780470125922.ch1)
T. Ziegler, A. Rauk, Inorg. Chem. 18 (1979) 1558 (https://doi.org/10.1021/ic50196a034)
K. B. Wiberg, Tetrahedron. 24 (1968) 1083 (https://doi.org/10.1016/0040-4020(68)88057-3)
M. Kohout, Program DGrid, version 4.3, 2008.
Chemcraft, Release 1.4, _http://www.chemcraftprog.com/>.
J. Kruszewski, T. M. Krygowski, Tetrahedron. Lett. 13 (1972) 3839 (https://doi.org/10.1016/S0040-4039(01)94175-9)
T. M. Krygowski, J. Chem. Inf. Comp. Sci. 33 (1993) 70 (https://doi.org/10.1021/ci00011a011)
E. Matito, M. Duran, M. Sola, J. Chem. Phys. 122 (2005). (https://doi.org/10.1063/1.1824895)
I. Petitpas, A. A. Bhattacharya, S. Twine, M. East, S. Curry, J. Biol. Chem. 276 (2001) 22804 (10.1074/jbc.M100575200)
Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017. Dassault Systèmes, San Diego, CA, USA, 2017.
G. M. Morris, R. Huey, W. Lindstrom et al, J. Comput. Chem. 30 (2009) 2785 (10.1002/jcc.21256)
O. Trott, A. J. Olson, J. Comput. Chem. 31 (2010) 455 (10.1002/jcc.21334)
W. J. Hehre, R. F. Stewart, J. A. Pople, J. Chem. Phys. 51 (1969) 2657 (https://doi.org/10.1063/1.1672392)
N. Kerru, L. Gummidi, S. V. H. S. Bhaskaruni, S. N. Maddila, P. Singh & B. S. Jonnalagadda, Scientific Reports. 9 (2019) 1 (https://doi.org/10.1038/s41598-019-55793-5)
P. Su, Z. Chen, W. Wu, Chemical Physics Letters. 635 (2015) 250 ((https://doi.org/10.1016/j.cplett.2015.06.078)
K. Shyan, A. Nowroozi, Structural Chemistry. 27 (2016) 1769 (https://doi.org/10.1007/s11224-016-0796-8)
S. J. Grabowski, Mol. Struct. 553 (2000) 151 (https://doi.org/10.1016/S0022-2860(00)00576-7)
E. D. Glendening, A. E. Reed, J. E. Carpenter, F. Weinhold, NBO, Version 3.1.
S. J. Grabowski, Phys. Chem. 102 (2006) 131 (https://doi.org/10.1039/B417200K)
S. Emamian, S. F. Tayyari, J. Chem. Sci. 125 (2013) 939 (https://doi.org/10.1007/s12039-013-0466-y)
G. Mahmoudzadeh, Int. J. New. Chem. 8 (2021) 277 (https://doi.org/10.22034/ijnc.2020.122797.1101)
J. D. Pedelacq, S. Cabantous, T. Tran, T. C. Terwilliger, and G. S. Waldo, Nat. Biotechnol. 24 (2006) 79 (10.1038/nbt1172)
P. V. R. Schleyer, Chem. Rev. 101 (2001) 1115 (https://doi.org/10.1021/cr0103221)
M. K. Cyrañski, Z. Czarnocki, G. Häfelinger, A. R. Katritzky, Tetrahedron. 56 (2000) 1783 (https://doi.org/10.1016/S0040-4020(99)00979-5)
J. Poater, M. Duran, M. Solà, B. Silvi, Chem. Rev. 105 (2005) 3911 (https://doi.org/10.1021/cr030085x)
C. A. Lipinski, F. Lombardo, B. W. Dominy & P. Feeney, J. Adv. Drug. Deliv. Rev. 46 (2001) 3 (10.1016/s0169-409x(00)00129-0)