Laboratory data clustering in defining population cohorts: Case study on metabolic indicators Scientific paper
Main Article Content
Abstract
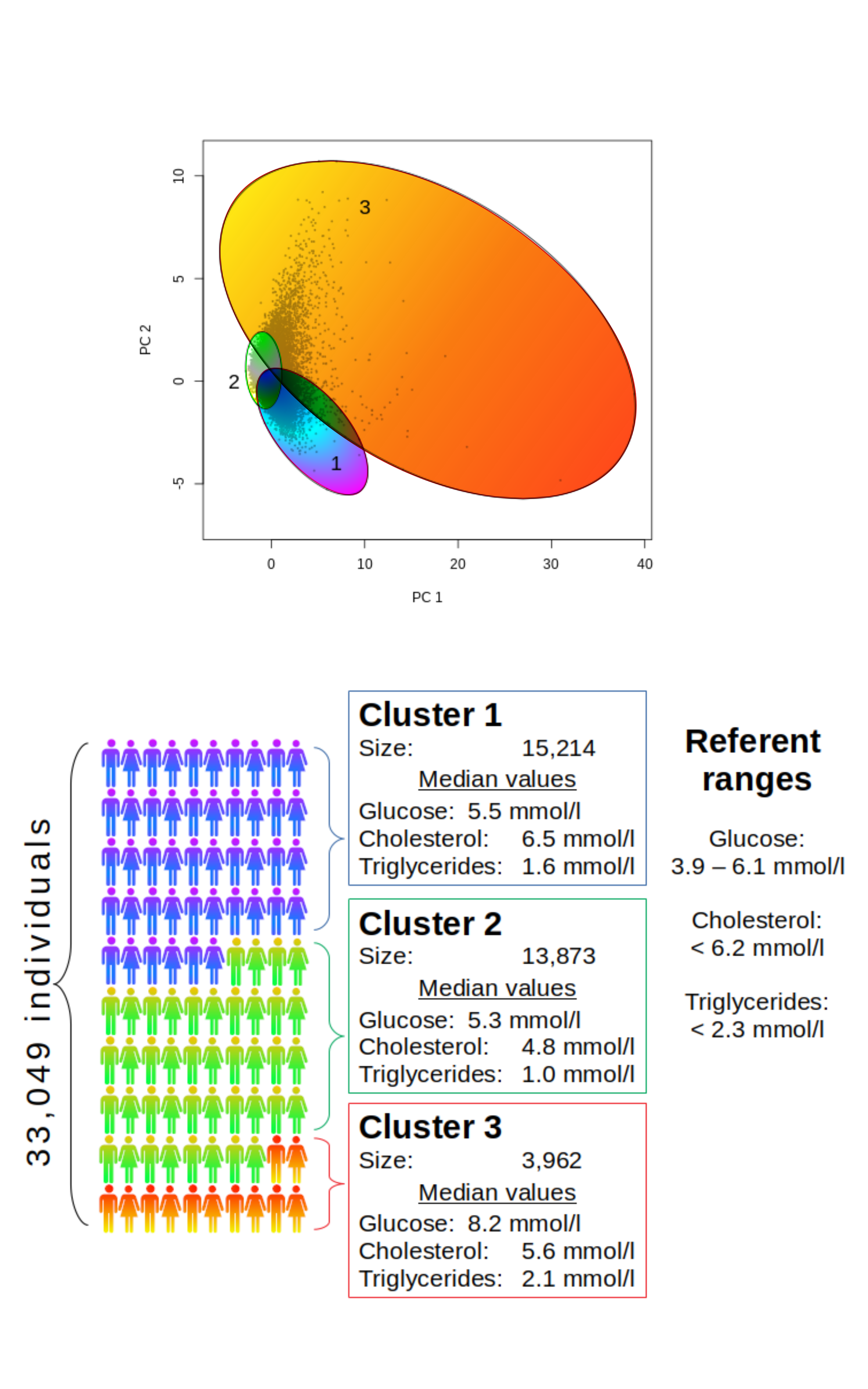
The knowledge on the general population health is important for creating public policies and organization of medical services. However, personal data are often limited, and mathematical models are employed to achieve a general overview. Cluster analysis was used in this study to assess general trends in population health based on laboratory data. Metabolic indicators were chosen to test the model and define population cohorts. Data on blood analysis of 33,049 persons, namely the concentrations of glucose, total cholesterol and triglycerides, were collected in a public health laboratory and used to define metabolic cohorts employing computational data clustering (CLARA method). The population was shown to be distributed in 3 clusters: persons with hypercholesterolemia with or without changes in the concentration of triglycerides or glucose, persons with reference or close to reference concentrations of all three analytes and persons with predominantly elevated all three parameters. Clustering of biochemical data, thus, is a useful statistical tool in defining population groups in respect to certain health aspect.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Ministarstvo Prosvete, Nauke i Tehnološkog Razvoja
Grant numbers 451-03-9/2021-14/
References
N. Sauvageot, A. Schritz, S. Leite, A. Alkerwi, S. Stranges, F. Zannad, S. Streel, A. Hoge, A.-F. Donneau, A. Albert, M. Guillaumeet, Nutr. J. 16 (2017) 4 (https://doi.org/10.1186/s12937-017-0226-9)
L. Kaufman, P. J. Rousseeuw, Finding groups in data: An introduction to cluster analysis, Wiley, New York, 1990 (ISBN 978-0-471-73578-6)
C. W. Hu, H. Li, A. A. Qutub, BMC Bioinform. 19 (2018) 19 (https://doi.org/10.1186/s12859-018-2022-8)
K. G. Parhofer, Diab. Metab. J. 39 (2015) 353 (https://doi.org/10.4093/dmj.2015.39.5.353)
G. H. Tomkin, D. Owens, Diab. Metab. Syndr. Obes. Targ. Ther. 10 (2017) 333 (https://doi.org/10.2147/DMSO.S115855)
J. Jensen, P. I. Rustad, A. J. Kolnes, Y. C. Lai, Front. Physiol. 2 (2011) 112 (https://doi.org/10.3389/fphys.2011.00112)
M. P. Czech, M. Tencerova, D. J. Pedersen, M. Aouadi, Diabetologia 56 (2013) 949 (https://doi.org/10.1007/s00125-013-2869-1)
C. J. Toro-Huamanchumo, D. Urrunaga-Pastor, M. Guarnizo-Poma, H. Lazaro-Alcantara, S. Paico-Palacios, B. Pantoja-Torres, V. Del Carmen Ranilla-Seguin, V. A. Benites-Zap¬ata, Diab. Metab. Syndr. 13 (2019) 272 (https://doi.org/10.1016/j.dsx.2018.09.010)
S. Huptas, H. C. Geiss, C. Otto, K. G. Parhofer, Am. J. Cardiol. 98 (2006) 66 (https://doi.org/10.1016/j.amjcard.2006.01.055)
H. Cederberg, A. Stancakova, N. Yaluri, S. Modi, J. Kuusisto, M. Laakso, Diabetologia 58 (2015) 1109 (https://doi.org/10.1007/s00125-015-3528-5)
S. B. Lee, M. K. Kim, S. Kang, K. Park, J. H. Kim, S. J. Baik, J. S. Nam, C. W. Ahn, J. S. Park, Endocrinol. Metab. 34 (2019) 179 (https://doi.org/10.3803/EnM.2019.34.2.179)
A. Kassambara, Practical guide to cluster analysis in R Unsupervised machine learning, STHDA, 2017 (www.sthada.com)
J. Krithikadatta, Conserv. Dent. 17 (2014) 96 (https://doi.org/10.4103/0972-0707.124171)
I. T. Jolliffe, Principal component analysis, Springer, New York, 2002 (https://doi.org/10.1007/b98835)
I. T. Jolliffe, J. Cadima, Phil. Trans., A 374 (2016) 20150202 (https://doi.org/10.1098/rsta.2015.0202)
F. Abbasi, G. M. Reaven, Metab. Clin. Exp. 60 (2011) 60 (https://doi.org/10.1016/j.metabol.2011.04.006)
L. E. Simental-Mendia, G. Hernandez-Ronquillo, R. Gomez-Diaz, M. Rodriguez-Moran, F. Guerrero-Romero, Pediatr. Res. 82 (2017) 920 (https://doi.org/10.1038/pr.2017.187)
T. Du, G. Yuan, M. Zhang, X. Zhou, X. Sun, X. Yu, Cardiovasc. Diabetol. 13 (2014) 146 (https://doi.org/10.1186/s12933-014-0146-3)
F. Guerrero-Romero, R. Villalobos-Molina, J. R. Jimenez-Flores, L. E. Simental-Mendia, R. Mendez-Cruz, M. Murguia-Romero, M. Rodríguez-Morán, Arch. Med. Res. 47 (2016) 382 (https://doi.org/10.1016/j.arcmed.2016.08.012)
J. Salazar, V. Bermudez, M. Calvo, L. C. Olivar, E. Luzardo, C. Navarro, H. Mencia, M. Martinez, J. Rivas-Rios, S. Wilches-Duran, M. Cerda, M. Graterol, R. Graterol, C. Garicano, J. Hernandez, J. Rojas, F1000Res. 6 (2017) 1337 (https://doi.org/10.12688/f1000research.12170.3)
L. E. Simental-Mendia, M. Rodriguez-Moran, F. Guerrero-Romero, Metab. Syndr. Rel. Disord. 6 (2008) 299 (https://doi.org/10.1089/met.2008.0034).