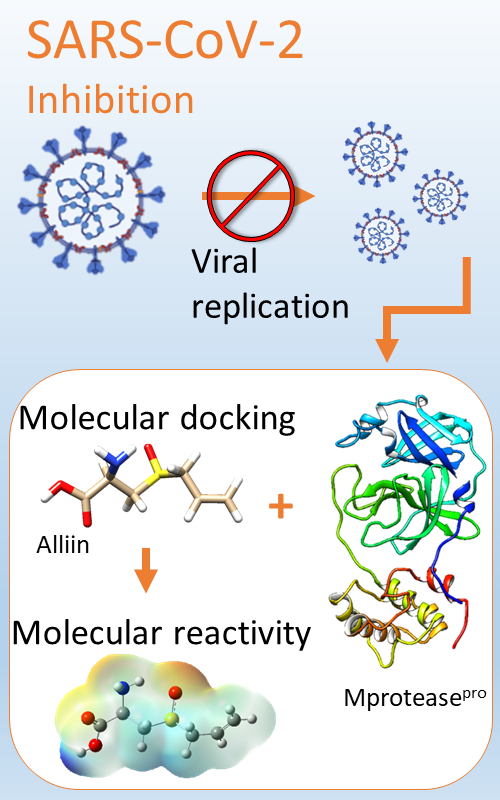
A Chemical reactivity of alliin and its molecular interactions with the M proteasepro of SARS-CoV-2.
Main Article Content
Abstract
In the present work a computational study of the chemical reactivity of alliin at the X/DGDZVP level of theory (where X=B3LYP, M06, M06L and wB97XD) was performed. The distribution of active sites on alliin was determined by evaluating the Fukui function. For electrophilic attacks, the more reactive sites are on the carbon atoms of the prop-2-ene moiety. The more active sites for nucleophilic attacks are located on the thioether group. In the case of free radical attacks, the more reactive sites are on the carbonyl, thioether and prop-2-ene moieties. Additionally, the molecular docking study revealed that, alliin is able to dock to the Mproteasepro of SARS-CoV-2 through interactions with the catalytic CYS145-HSD164 dyad via Van der Waals interactions, with MET49 with interactions alkyl-type ions and with PHE140 by hydrogen bonds. Also, the molecular dynamic study indicates that alliin remains in the pocket site. Last result suggests that this molecule is a potential candidate for further in vitro evaluation as a drug for the treatment of the major protease-based SARS-CoV-2 virus.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Consejo Nacional de Ciencia y Tecnología
Grant numbers CB2015-257823;CONACyT 123732
References
B. Hughes, B. Murray, J. North, L. Lawson, Planta Med 55 (1989) 114–114. (https://www.thieme-connect.com/products/ejournals/abstract/10.1055/s-2006-961894)
S. Quintero-Fabián, D. Ortuño-Sahagún, M. Vázquez-Carrera, R. I. López-Roa, Mediators Inflamm. 2013 (2013). (https://doi.org/10.1155/2013/381815)
M. G. Jones, J. Hughes, A. Tregova, J. Milne, A. B. Tomsett, H. A. Collin, J. Exp. Bot. 55 (2004) 1903–1918. (https://doi.org/10.1093/jxb/erh138)
C. Jacob, A. Anwar, Physiol. Plant. 133 (2008) 469–480. (https://doi.org/10.1111/j.1399-3054.2008.01080.x)
N. D. Weber, D. O. Andersen, J. A. North, B. K. Murray, L. D. Lawson, B. G. Hughes, Planta Med. 58 (1992) 417–423. (https://www.thieme-connect.com/products/ejournals/abstract/10.1055/s-2006-961504)
A. Helen, K. Krishnakumar, P. L. Vijayammal, K. T. Augusti, Pharmacology 67 (2003) 113–117. (https://doi.org/10.1159/000067796)
D. A. Hanoush, A. H. Al-Auqaili, M. Mansour, A. Ghosh, Trop. J. Nat. Prod. Res. 6 (2022) 1233–1240. (http://www.doi.org/10.26538/tjnpr/v1i4.5)
J. M. Casasnovas, T. A. Springer, J. Biol. Chem. 270 (1995) 13216–13224. (https://doi.org/10.1074/jbc.270.22.13216)
P. Gale, Microb. Risk Anal. 21 (2022) 100198. (https://doi.org/10.1016/j.mran.2021.100198)
M. E. Popovic, Microbiol. Res. 270 (2023) 127337. (https://doi.org/10.1016/j.micres.2023.127337)
K. Rajagopal, G. Byran, S. Jupudi, R. Vadivelan, Int. J. Heal. Allied Sci. 9 (2020) 43–50. (http://www.doi.org/10.4103/ijhas.IJHAS_55_20)
W. Lopez-Orozco, L. H. Mendoza-Huizar, G. A. Álvarez-Romero, J. de J. M. Torres-Valencia, Pädi Boletín Científico Ciencias Básicas e Ing. Del ICBI 10 (2023) 126–130. (https://repository.uaeh.edu.mx/revistas/index.php/icbi)
M. B. M. Spera, F. A. Quintão, D. K. D. Ferraresi, W. R. Lustri, A. Magalhães, A. L. B. Formiga, P. P. Corbi, Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 78 (2011) 313–318. (https://doi.org/10.1016/j.saa.2010.10.012)
P. Geerlings, F. De Proft, W. Langenaeker, Chem. Rev. 103 (2003) 1793–1873. (https://doi.org/10.1021/cr990029p)
R. G. Pearson, J. Chem. Educ. 64 (1987) 561–567. (https://doi.org/10.1021/ed064p561)
R. G. Parr, R. A. Donnelly, M. Levy, W. E. Palke, J. Chem. Phys. 68 (1977) 3801–3807. (https://doi.org/10.1063/1.436185)
R. G. Parr, R. G. Pearson, J. Am. Chem. Soc. 105 (1983) 7512–7516. (https://doi.org/10.1021/ja00364a005)
R. G. Parr, W. Yang, J. Am. Chem. Soc. 106 (1984) 4049–4050. (https://doi.org/10.1021/ja00326a036)
R. G. Parr, Horizons Quantum Chem. (1980) 5–15. (https://link.springer.com/book/10.1007/978-94-009-9027-2)
W. Yang, W. J. Mortier, J. Am. Chem. Soc. 108 (1986) 5708–5711. (https://doi.org/10.1021/ja00279a008)
N. Godbout, D. R. Salahub, J. Andzelm, E. Wimmer, Can. J. Chem. 70 (2011) 560–571. (https://doi.org/10.1139/v92-079)
K. Raghavachari, Theor. Chem. Accounts 103 (2000) 361–363. (https://link.springer.com/article/10.1007/s002149900065)
Y. Zhao, D. G. Truhlar, Theor. Chem. Acc. 120 (2008) 215–241. (https://link.springer.com/article/10.1007/s00214-007-0310-x)
Y. Wang, X. Jin, H. S. Yu, D. G. Truhlar, X. He, Proc. Natl. Acad. Sci. U. S. A. 114 (2017) 8487–8492. (https://doi.org/10.1073/pnas.1705670114)
J. Da Chai, M. Head-Gordon, Phys. Chem. Chem. Phys. 10 (2008) 6615–6620. (https://doi.org/10.1039/B810189B)
S. Miertuš, E. Scrocco, J. Tomasi, Chem. Phys. 55 (1981) 117–129. (https://doi.org/10.1016/0301-0104(81)85090-2)
Gaussian 09, Revision A.01, Gaussian, Inc., Wallingford, CT, 2009. (https://gaussian.com/g09citation/)
Gaussview Rev. 3.09, Windows version. Gaussian Inc., Pittsburgh, PA. (https://gaussian.com/508_gvw/)
A. Allouche, J. Comput. Chem. 32 (2012) 174–182. (https://doi.org/10.1002/jcc.21600)
T. Lu, F. Chen, J. Comput. Chem. 33 (2012) 580–592. (https://doi.org/10.1002/jcc.22885)
Jorge-Finnigan, A.; Brasil, S.; Underhaug, J.; Ruíz-Sala, P.; Merinero, B.; Banerjee, R.; Desviat, L. R.; Ugarte, M.; Martinez, A.; Pérez, B. Pharmacological chaperones as a potential therapeutic option in methylmalonic aciduria cblB type. Hum. Mol. Genet. 18(2013). (https://doi.org/10.1093/hmg/ddt217)
E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin, J. Comput. Chem. 25 (2004) 1605–1612. (https://doi.org/10.1002/jcc.20084)
BIOVIA, Dassault Systèmes, Discovery Studio Visualiser 2019, San Diego: Dassault Systèmes, 2019. (https://discover.3ds.com/discovery-studio-visualizer-download)
J. L. Gázquez, J. Phys. Chem. A 101 (1997) 4591–4593. (https://www.scielo.org.mx/scielo.php?script=sci_arttext&pid=S1870-249X2008000100002)
P. K. Chattaraj, (2009) 576. (https://books.google.com/books/about/Chemical_Reactivity_Theory.html?hl=es&id=n8JBNvF_2KAC)
F. L. Hirshfeld, Theor. Chim. Acta 44 (1977) 129–138. (https://link.springer.com/article/10.1007/BF00549096)
E. R. Johnson, S. Keinan, P. Mori-Sánchez, J. Contreras-García, A. J. Cohen, W. Yang, J. Am. Chem. Soc. 132 (2010) 6498–6506. (https://doi.org/10.1021/ja100936w)
M. Chebaibi, D. Bousta, R. F. B. Goncalves, H. Hoummani, S. Achour, Res. Sq.1 (2021). (https:// https://doi.org/10.21203/rs.3.rs-679827/v1)
Z. Jin, X. Du, Y. Xu, Y. Deng, M. Liu, Y. Zhao, Nature 582(2020) 289-293. (https://doi.org/10.1038/s41586-020-2223-y)
A. Castro-Alvarez, A. M. Costa, J. Vilarrasa, Molecules 22 (2017). (https://doi.org/10.3390/molecules22010136).