On the importance of π–π interactions in structural stability of phycocyanins Scientific paper
Main Article Content
Abstract
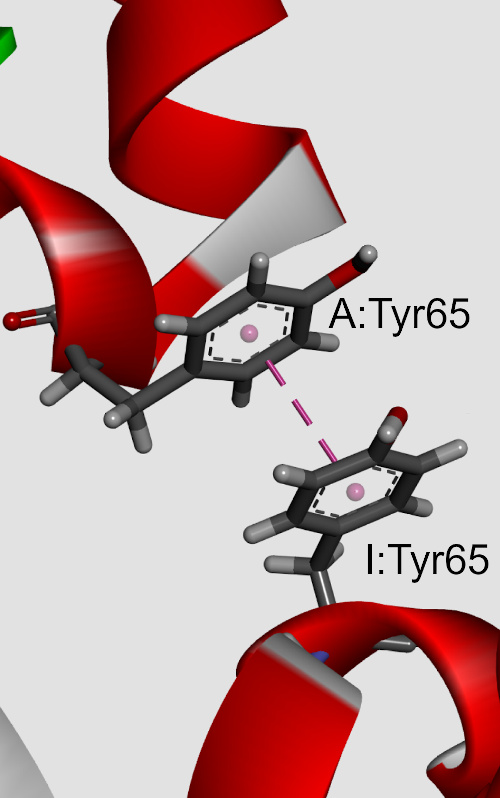
The influences of π-π interactions in phycocyanin proteins and their environmental preferences were analyzed. The observations indicate that the majority of the aromatic residues in phycocyanin proteins are involved in π-π interactions. Phenylalanine (Phe) and tyrosine (Tyr) residues were found to be involved in π–π interactions much more frequently than tryptophan (Trp) or histidine (His). Similarly, the Phe-Phe and Tyr-Tyr π–π interacting paiThe influences of π-π interactions in phycocyanin proteins and their environmental preferences were analyzed. The observations indicate that the majority of the aromatic residues in phycocyanin proteins are involved in π-π interactions. Phenylalanine (Phe) and tyrosine (Tyr) residues were found to be involved in π–π interactions much more frequently than tryptophan (Trp) or histidine (His). Similarly, the Phe-Phe and Tyr-Tyr π–π interacting pair had the highest frequency of occurrence. In addition to π-π interactions, the aromatic residues also form π-networks in phycocyanins. The π–π interactions are most favourable at the pair distance range of 5.5–7 Å, with a clear preference for T-shaped ring arrangements. Using ab initio calculations, we observed that most of the π-π interactions possess energy from 0 to -10 kJ mol-1. Stabilization centres for these proteins showed that all residues found in π-π interactions are important in locating one or more such centres. π-π interacting residues are evolutionary conserved. The results obtained from this study will be beneficial in further understanding the structural stability and eventual development of protein engineering of phycocyanins.r had the highest frequency of occurrence. In addition to π-π interactions, the aromatic residues also form π-networks in phycocyanins. The π–π interactions are most favourable at the pair distance range of 5.5–7 Å, with a clear preference for T-shaped ring arrangements. Using ab initio calculations, we observed that most of the π-π interactions possess energy from 0 to -10 kJ mol-1. Stabilization centres for these proteins showed that all residues found in π-π interactions are important in locating one or more such centres. π-π interacting residues are evolutionary conserved. The results obtained from this study will be beneficial in further understanding the structural stability and eventual development of protein engineering of phycocyanins.
Downloads
Metrics
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.

Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution license 4.0 that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
Funding data
-
Ministarstvo Prosvete, Nauke i Tehnološkog Razvoja
Grant numbers 451-03-47/2023-01/ /200026 -
Ministarstvo Prosvete, Nauke i Tehnološkog Razvoja
Grant numbers 451-03-47/2023-01/200168
References
N. Tandeau de Marsac, Photosynth. Res. 76 (2003) 193 (https://doi.org/10.1023/A:1024954911473)
P. Falkowski, R. J. Scholes, E. Boyle, J. Canadell, D. Canfield, J. Elser, N. Gruber, K. Hibbard, P. Hogberg, S. Linder, F. T. Mackenzie, B. Moore, III, T. Pedersen, Y. Rosenthal, S. Seitzinger, V. Smetacek, W. Steffen, Science 290 (2000) 291 (https://doi.org/10.1126/science.290.5490.291)
V. K. Kannaujiya, D. Kumar, V. Singh, R. P. Sinha, in Natural Bioactive Compounds, R. Sinha, D. P. Häder, Eds., Academic Press, New York, 2021, pp. 57––81 ISBN: 0128206594
M. G. de Morais, D. da Fontoura Prates, J. B. Moreira, J. H. Duarte, J. A. V. Costa, Ind. Biotechnol. 14 (2018) 30 (https://doi.org/10.1089/ind.2017.0009)
McGregor, M. Klartag, L. David, N. Adir, J. Mol. Biol. 384 (2008) 406 (https://doi.org/10.1016/j.jmb.2008.09.018)
D. Andersson, B. K. Mishra, N. Forsgren, F. Ekström, A. Linusson, J. Phys. Chem., B 124 (2020) 6529 (https://doi.org/10.1021/acs.jpcb.0c03778)
E. Lanzarotti, L. A. Defelipe, M. A. Marti, A. n. G. Turjanski, J. Cheminform. 12 (2020) 30 (https://doi.org/10.1186/s13321-020-00437-4)
K. S. Chatterjee, R. Das, J. Biol. Chem. 297 (2021) (https://doi.org/10.1016/j.jbc.2021.100970)
H. B. Gray, J. R. Winkler, Chem. Sci. 12 (2021) 13988 (https://doi.org/10.1039/D1SC04286F)
Z. Y. Yan, X. J. Xu, L. Fang, C. Geng, Y. P. Tian, X. D. Li, Phytopathology Res. 3 (2021) 10 (https://doi.org/10.1186/s42483-021-00088-9)
M. O. Sinnokrot, C. D. Sherrill, J. Am. Chem. Soc. 126 (2004) 7690 (https://doi.org/10.1021/ja049434a)
R. Bhattacharyya, U. Samanta, P. Chakrabarti, Protein Eng. Des. Sel. 15 (2002) 91 (https://doi.org/10.1093/protein/15.2.91)
N. Kannan, S. Vishveshwara, Protein Eng. Des. Sel. 13 (2000) 753 (https://doi.org/10.1093/protein/13.11.753)
S. Tsuzuki, K. Honda, T. Uchimaru, M. Mikami, K. Tanabe, J. Am. Chem. Soc. 124 (2002) 104 (https://doi.org/10.1021/ja0105212)
V. Morozov, K. M. S. Misura, K. Tsemekhman, D. Baker, J. Phys. Chem. B. 108 (2004) 8489 (https://doi.org/10.1021/jp037711e)
E. Lanzarotti, R. R. Biekofsky, D. o. A. Estrin, M. A. Marti, A. n. G. Turjanski, J. Chem. Inf. Model. 51 (2011) 1623 (https://doi.org/10.1021/ci200062e)
L. M. Breberina, M. V. Zlatović, M. R. Nikolić, S. Đ. Stojanović, Mol. Inform. 38 (2019) e1800145 (https://doi.org/10.1002/minf.201800145)
L. M. Breberina, M. R. Nikolić, S. Đ. Stojanović, M. V. Zlatović, Comput. Biol. Chem. 100 (2022) 107752 (https://doi.org/10.1016/j.compbiolchem.2022.107752)
P. W. Rose, B. Beran, C. Bi, W. F. Bluhm, D. Dimitropoulos, D. S. Goodsell, A. Prlic, M. Quesada, G. B. Quinn, J. D. Westbrook, J. Young, B. Yukich, C. Zardecki, H. M. Berman, P. E. Bourne, Nucleic Acids Res. 39 (2011) D392 (https://doi.org/10.1093/nar/gkq1021)
G. Murzin, S. E. Brenner, T. Hubbard, C. Chothia, J. Mol. Biol. 247 (1995) 536 (https://doi.org/10.1016/S0022-2836(05)80134-2)
J. M. Word, S. C. Lovell, J. S. Richardson, D. C. Richardson, J. Mol. Biol. 285 (1999) 1735 (https://doi.org/10.1006/jmbi.1998.2401)
Discovery Studio Visualizer, release 2020, Accelrys Software Inc., San Diego, CA, 2020
G. B. McGaughey, M. Gagné, A. K. Rappé, J. Biol. Chem. 273 (1998) 15458 (https://doi.org/10.1074/jbc.273.25.15458)
V. R. Ribić, S. Đ. Stojanović, M. V. Zlatović, Int. J. Biol. Macromol. 106 (2018) 559 (https://doi.org/10.1016/j.ijbiomac.2017.08.050)
J. Hostaš, D. Jakubec, R. A. Laskowski, R. Gnanasekaran, J. Řezáč, J. Vondrášek, P. Hobza, J. Chem. Theory Comput. 11 (2015) 4086 (https://doi.org/10.1021/acs.jctc.5b00398)
D. Bochevarov, E. Harder, T. F. Hughes, J. R. Greenwood, D. A. Braden, D. M. Philipp, D. Rinaldo, M. D. Halls, J. Zhang, R. A. Friesner, Int. J. Quantum Chem. 113 (2013) 2110 (https://doi.org/10.1002/qua.24481)
T. H. Dunning, J. Chem. Phys. 90 (1989) 1007 https://doi.org/10.1063/1.456153
T. Clark, J. Chandrasekhar, G. n. W. Spitznagel, P. V. R. Schleyer, J. Comput. Chem. 4 (1983) 294 (https://doi.org/10.1002/jcc.540040303)
K. E. Riley, J. A. Platts, J. Řezáč, P. Hobza, J. G. Hill, J Phys. Chem., A 116 (2012) 4159 (https://doi.org/10.1021/jp211997b)
G. J. Jones, A. Robertazzi, J. A. Platts, J. Phys. Chem., B 117 (2013) 3315 (https://doi.org/10.1021/jp400345s)
S. Saebø, W. Tong, P. Pulay, J. Chem. Phys. 98 (1993) 2170 (https://doi.org/10.1063/1.464195)
Reyes, L. Fomina, L. Rumsh, S. Fomine, Int. J. Quantum Chem. 104 (2005) 335 (https://doi.org/10.1002/qua.20558)
R. M. Balabin, J. Chem. Phys. 132 (2010) 231101 (https://doi.org/10.1063/1.3442466)
Y. Deng, B. t. Roux, J. Phys. Chem., B 113 (2009) 2234 https://doi.org/10.1021/jp807701h
J. C. Gumbart, B. t. Roux, C. Chipot, J. Chem. Theory Comput. 9 (2013) 794 https://doi.org/10.1021/ct3008099
J. Černý, P. Hobza, Phys. Chem. Chem. Phys. 9 (2007) 5291 https://doi.org/10.1039/B704781A
Z. Dosztányi, A. Fiser, I. Simon, J. Mol. Biol. 272 (1997) 597 (https://doi.org/10.1006/jmbi.1997.1242)
Z. Dosztányi, C. Magyar, G. Tusnady, I. Simon, Bioinformatics 19 (2003) 899 (https://doi.org/10.1093/bioinformatics/btg110)
H. Ashkenazy, E. Erez, E. Martz, T. Pupko, N. Ben-Tal, Nucleic Acids Res. 38 (2010) W529 (https://doi.org/10.1093/nar/gkq399)
B. Boeckmann, A. Bairoch, R. Apweiler, M. C. Blatter, A. Estreicher, E. Gasteiger, M. J. Martin, K. Michoud, C. O'Donovan, I. Phan, S. Pilbout, M. Schneider, Nucleic Acids Res. 31 (2003) 365 (https://doi.org/10.1093/nar/gkg095)
C. A. Hunter, J. Singh, J. M. Thornton, J. Mol. Biol. 218 (1991) 837 (https://doi.org/10.1016/0022-2836(91)90271-7)
F. Cozzi, M. Cinquini, R. Annunziata, T. Dwyer, J. S. Siegel, J. Am. Chem. Soc. 114 (1992) 5729 (https://doi.org/10.1021/ja00040a036)
S. Mahadevi, G. N. Sastry, Chem. Rev. 116 (2016) 2775 (https://doi.org/10.1021/cr500344e)
Ma, T. Elkayam, H. Wolfson, R. Nussinov, Proc. Natl. Acad. Sci. USA 100 (2003) 5772 (https://doi.org/10.1073/pnas.1030237100)
J. B. Mitchell, R. A. Laskowski, J. M. Thornton, Proteins 29 (1997) 370 (https://doi.org/10.1002/(SICI)1097-0134(199711)29:3%3C370::AID-PROT10%3E3.0.CO;2-K)
E. G. Hohenstein, C. D. Sherrill, J. Phys. Chem., A 113 (2009) 878 (https://doi.org/10.1021/jp809062x)
P. Chakrabarti, R. Bhattacharyya, Prog. Biophys. Mol. Biol. 95 (2007) 83 (https://doi.org/10.1016/j.pbiomolbio.2007.03.016)
B. P. Dimitrijević, S. Z. Borozan, S. Đ. Stojanović, RSC Adv. 2 (2012) 12963 (https://doi.org/10.1039/C2RA21937A)
P. A. Maury, D. N. Reinhoudt, J. Huskens, Curr. Opin. Colloid Interface Sci. 13 (2008) 74 (https://doi.org/10.1016/j.cocis.2007.08.013)
M. Landau, I. Mayrose, Y. Rosenberg, F. Glaser, E. Martz, T. Pupko, N. Ben-Tal, Nucleic Acids Res. 33 (2005) W299-W302 (https://doi.org/10.1093/nar/gki370).